EBCAs biology in plants, pests and pathogens interactions
Task 3.1 – Molecular biology of EBCAs.
A gene expression study on Gran Enana plants artificially infected by Foc TR4, with or without addition of P. chlamydosporiaDSM 26985 is in course at CNR, in a quarantine conditioned greenhouse. Aim is checking if root colonization by P. chlamydosporiaelicits expression of defense genes such as PAL 5, PIN II, PR1and LOX by means of RT-PCR, at different days post-infection. The greenhouse plants have also been used for whole transcriptome analysis. A preliminary trascriptomic study has also been carried out through in vitroinoculation of Gran Enana roots with Pratylenchus goodeyispecimens extracted from Tenerife soil (50 nem per plantlet), with or without inoculation with P. chlamydosporia. Work is in progress.
Task 3.2 – Root biology with endophytic EBCAs in PPN and banana / enset root interactions
Subtask: 3.2.1. Microscopy analysis of the interaction of P. fluorescens PICF7 with PPN, and PPN / enset interactions.
Many root-associated Pseudomonas spp. strains are known as beneficial to plants either because of their plant-growth promotion effect or their potential as biolcontrol agents. Some of them may also establish endophytically, and have been identified and/or isolated from a wide range of plant species. Pseudomonas fluorescens PICF7 has been characterized by CSIC as an effective biological control agent of Verticillium wilt of olive. Moreover, this bacterium is able to colonize and persist on roots of olive (the host from which is was originally isolated), wheat, barley, sunflower and Arabidopsis thaliana. Evidence of root endophytic colonization has been obtained except for the last two species. Therefore, CSIC stated two independent pilot bioassays in order to check whether a fluorescently-tagged derivative of P. fluorescens PICF7 was able to colonize banana roots endophytically.
For a first bioassay, banana cv. Pequeña Enana plants were uprooted from the original substrate, their roots thoroughly washed in tap water without wounding and dipped for 15 min in a bacterial suspension (1・108cfuml−1) of P. fluorescens strain PICF7 (pMP4655). For the control treatment, other fifteen plants were treated similarly except that the root systems were dipped in 10 mM MgSO4·7H2O. Plantsinoculation in a second trial was performed by adding 15 ml of a suspension of bacterial cells (1・108cfuml−1in sterile MgSO4·7H2O 10 mM) per pot. For the control treatment, other fifteen plants were treated similarly except that the root systems were drenched with 10 mM MgSO4 · 7H2O. After inoculation, the plants were grown in controlled chamber with 28 ± 1 °C, 75–80% RH, 14-h photoperiod of fluorescent light. Several segments representative of the whole root system were analysed on a confocal laser microscope over 8 days after bacterial inoculation.Two non-bacterized, control plants were also sampled at the first and eighth days to check for plant tissue autofluorescence, possible fluorescent native bacteria or cross contamination. So far, no consistent microscopy evidence showed that strain PICF7 efficiently colonized banana roots. Further trials will be conducted in order to verify whether strain PICF7 is able to colonize banana roots and check its effects on PPN and related antagonists.
Subtask: 3.2.2 – Rhizosphere competence.
UA devised three systems for analysing interactions of inoculation withP. chlamydosporia on banana host plants. The isolate of P. chlamydosporiaPc 123 (ATCC n. MYA-4875; CRCT n. 20929) was tested on in vitrocommercial plants of cv. Pequeña Enana, using 30-day-old in vitro plantlets on agar medium or 6-7 week-old in vitro derivedplants, grown in planting trays, in the greenhouse. The banana-EBCAs interaction schemes devised were the following:
- axenic bioassays in magenta boxes for 1-7 days.
- experiments in 200 ml cups, in growth chambers for up to 30 days
- experiments in 1L/3.5 L pot, in the greenhouse for 2.5 months.
Thirty-day-old in vitro banana plantlets (both controls and EBCA inoculated) were planted in cups with sterilized peat moistened with 1/10 Gamborg’s nutrient solution, keptin growth chambers at 24ºC and 60% RH for 10, 20 and 30 days. The effect of P. chlamydosporia inoculation on banana plantlets was then estimated, recording root, corm and leaf growth parameters. Three treatments to inoculate the fungus were tested.
Treatment 1 involved plant inoculation with P. chlamydosporiamycelium, placing eight cores from the edge of a 21-day-old colony in a planting hole in the cups. Treatment 2 consisted of mycelium inoculation by direct root contact using a 5 days old P. chlamydosporiacolony. In early tests banana plantlet roots were placed in contact with P. chlamydosporiacolonies on agar medium growing in petri dishes. This method appeared unsuitable as the agar dried-up after five days. As an alternative, colonies for plantlet inoculation in polycarbonate magenta boxes were used with 60 ml of poured CMA per box, autoclaved for 20 min at 121ºC. The CMA magenta were inoculated with P. chlamydosporiamycelial cores, and incubated for 15 days at 24ºC. The plantlets were inoculated by axenically placing roots in contact with a P. chlamydosporiacolony in a magenta for 5 days, and then planted in polystyrene cups with sterile peat.
Treatment 3 involved banana plantlet inoculation with 104propagules (conidia and chlamydospores) per g of substrate, obtained from fungus cultures grown on pre-hydrated rice grains. Flasks with inoculated rice were incubated in the dark at room temperature 3-4 weeks (Fig. 1). Twenty g of P. chlamydosporia-inoculated rice were shaken 1 min with 50 ml distilled water in sterile Falcon tubes. The resulting suspension was filtered and the fungusconidia and chlamydospores in the suspension were counted. Plant inoculation was performed by adding, close to the banana plantlets, a volume of suspension (ca. 500 microliters) to inoculate 104propagules per g of substrate (peat). Controls include sterilized peat in cups and uninoculated plantlets.

Ten plantlets were sampled after 10, 20 and 30 days. Root growth parameters were maximum root length (cm), fresh root weight (g) and the percent of total and endophytic colonization of the roots, using semi-selective media with antibiotics. Corm growth parameters were corm length (cm) and fresh and dry weight (g). The numbers of leaves per plant were counted, and maximum leaf length (cm) and fresh and dry leaf weight (g) were recorded.
Inoculations for greenhouse experiments. Experiments were carried out in a greenhouse at 24ºC and 60% RH, with 6-7 week old banana plants in 1 to 3.5 L pots. First experiment was performed initially in 1 L pots. The second replicate was set in 3.5 L pots. The main objective was to test methods for inoculating P. chlamydosporiain banana plants, and verify if the treatments were effective in the long term. Four methods of inoculation were developed. After two and a half months, parameters of the different parts of the banana plants were recorded, such as root, corm, pseudostem and leaves.
For transplants, it was necessary to fill the 1L/3.5L pots to 80% of their capacity with peat and then moisten it. Ahole was then made in the peat and a banana plant was planted in each pot. All plants were watered with an automated irrigation system.
For 1L pots, treatment 1 involved 4 agar and mycelium cores for each plant, arranged with roots in contact with the fungus. Treatment 2, the same as the previous one, was applied using 12 agar cores with P. chlamydosporia mycelium for each plant. Treatment 3 involved plant inoculation with 5000 conidia/chlamydospores (5.000 c/c) suspension per g of substrate. Treatment 4 was similar to the previous one, using 104conidia/chlamydospores. Controls included peat in the 1 L pot and banana plants only. For 3.5 L pots experiments only treatments 3 and 4 were applied, with a single or 3 times monthly inoculations.
Growth chamber experiments.All treatments with P. chlamydosporiaapplied to Pequeña Enana promoted root growth. However, the plantlets showed growth parameters significantly higher than control (uninoculated plants), with a maximal root length achieved when treated with conidial and chlamydospore suspensions. Similar results were found for the fresh root weight.
Surface sterilization of banana plantlet roots was initially performed using 0.5% sodium hypochlorite for estimating P. chlamydosporiaendophytic colonization. This concentration was increased to 1% since 0.5% caused positiveP. chlamydosporiagrowth in roots imprints (false positives for endophytic colonization). The fungus colonised endophyticaly banana plantlets roots in all treatments tested.
No significant difference between P. chlamydosporiatreatments and control was found. Treatment 1 (8 agar cores) increased corm length. After 20 days (treatment 3) P. chlamydosporiasignificantly increased corm weight of banana plantlets. Inoculation significantly increased leaf lengths at 10, 20 and 30 days after treatment 3 (inoculation with conidia and chlamydospores) and leaf fresh weight (especially 30 days after treatment 3). Similarly, the fungus significantly increased leaf dry weight 20 days after treatment 3 (conidia and chlamydospores). Treatments, however, did not affect significantly the number of leaves per plant, with a slight increase in plants of treatment 3 only.
Greenhouse assays. Greenhouse experiments were started using 1L pots using 20 day-old plants supplied by Cultesa (Tenerife, ES). In this experiments 4 treatments were implemented (inoculation with 4 P. chlamydosporia plugs, inoculation with 12 P. chlamydosporia plugs, both from a 21-day-old colony on CMA, inoculation with 5,000 and 10,000 c/c). Plants were incubated under greenhouse conditions for 75 days (2.5 months), measuring growth parameters (root, corm, pseudostem and leaves length and weight) at the end of the experiments. Results indicated that the fresh root weight of the banana plants was reduced when P. chlamydosporiamycelium was applied under this conditions. However, inoculation caused no significant differences in root length. For maximum leaves length, treatment 3 (5,000 c/c suspension) showed the best effect compared to control. However, no statistical difference was found between treatments and control, although differences were found among treatments.
Treatments with 4 and 12 cylinders of mycelium per plant reduced fresh leaf weight respect to the control, significantly for treatment with 12 plugs. Evaluating corm paramters, a large variability in length among samples was observed. Therefore, no significant difference between fungal treatments and controls was found. Conidia and chlamydospores slightly increased corm weight. In addition, no significant differences between treated plants and control were found for pseudostem length and weight. As this effect could be due to the smaller size of the pots used (plants could not develop rhizosphere system during the experiment) a larger pot size (3.5L) was used for subsequent experiments.
Colonization by P. chlamydosporiaof Pequeña Enana plants was scored under greenhouse conditions with the inoculation methods described above. For colonization, root fragments that showed occurrence of P. chlamydosporiawere scored. Roots were surface sterilised for endophytic colonization, whereas for total colonization they were only washed with sterile distilled water. Inoculation with conidia and chlamydospores increased endophytism and total root colonization in comparison to plants inoculated with mycelium (plugs treatments 4 cil, 12 cil). Pochonia chlamydosporiaalso significanly increased (p<0.05) pseudostem length and fresh weight, in treated plants.
Effect of plant substrate onP. chlamydosporia germination.Use of peat as banana plant growth substrate showed that it significantly reduced (p <0.05) germination of P. chlamydosporiaconidia and chlamydospores. The effect was maintained over time (for at least 48 h). A reduction was observed in both peat extracts sterilized by filtration and not filtered, suggesting an inhibition of germination that was independent from peat-associated microbiota. The active compound(s) present in the peat, inhibitory of P. chlamydosporiagermination, also appeared heat-resistant (withstanding steam sterilisation).
Subtask: 3.2.3 Soil receptivity
An assay is actually under evaluation at UA for the study of soil receptivity to selected P. chlamydosporia, based on a soil membrane technique.
Three independent pilot trials were conducted at IAS CSIC under non-gnotobiotic conditions in order to: (i) familiarize and optimize banana cultivation and ii) set up experimental conditions to assess PD symptoms development under controlled conditions. Banana plants (in vitropropagated seedlings, cv. Pequeña Enana, susceptible to PD, supplied by Cultesa) were first acclimated under controlled growth-chamber conditions (23 ± 1 °C, 75–80% RH, 14-h photoperiod) during fifteen days. After that, plants were transplanted to pots each containing ad hocprepared substrate (peat:sand:vermiculite, 75:12.5:12.5, v:v:v) without wounding roots. Then, plants were kept in a controlled growth chamber under the same conditions mentioned above.
In the first experiment (I), 60 (five-month-old) banana plants were challenged with three Fusarium oxysporumf. sp. cubense(Foc) strains from (CAV-050 and CAV-095 [STR4] and CAV-183 [R1 or R2], kindly provided by Prof. Viljoen, and one from Tenerife Island (3G [undetermined race], isolated from a banana plant showing PD symptoms during the first sampling round), by adding 150 ml per pot of a conidia suspension of each Foc isolate (1·106 conidial/ml). Non-inoculated plants (control) were only watered. Plants were grown in a controlled growth chamber with 28 ± 1 °C, 75–80% RH, 14-h photoperiod, with a random distribution.
In the second bioassay (II), 60 (6-month-old) banana plants were challenged with three isolates from Prof. Viljoen collection (CAV-050, CAV-095 and CAV-020 [STR4]) and two from Tenerife Island (2A and 3B, undetermined race).
Finally, a third trial (III) was conducted using 20 (one-month-old) banana plants to evaluate if disease symptoms developed in a similar way to those observed in older plants. Plants were transplanted twice before Foc inoculation with CAV-050 (STR4).
Banana plants grew well under controlled growth-chamber conditions (Fig. 2). Forty five days after inoculation, PD symptoms were observed in Foc-inoculated plants, in contrast to non-inoculated plants (Fig. 3 and 4). In experiment I, however, plants inoculated with the R1/R2 strain did not show Fusarium wilt symptoms since ‘Pequeña Enana’ cultivar is not susceptible to these races (Fig. 3C). Two months after inoculation, disease progress seems to stop, and non-inoculated and inoculated plants showed a very similar aspect. Symptoms of natural senescence were observed and Foc-inoculated plants recovered from the disease. Further pathogenicity tests will be performed in order to test different inoculum densities and inoculation methods.
New pathogenicity tests are under evaluation at CSIC with a different inoculation method, increasing pot size and modifying substrate (peat:sand:vermiculite, 1/3:1/3:1/3, v:v:v), to improve disease development observed in the previous assays (Fig. 5).




Subtask: 3.2.4 Identification of P. chlamydosporiagenes involved in plant growth promotion and PPN parasitism.
A new gene prediction of the P. chlamydosporia123 (Pc) genome was carried out in cooperation beteween UA and UNEXE, allowing the identification of novel genes. Transcriptomic studies on the fungus were carried out by UA and CNR, and sequencing results are being analysed.
Task 3.3 – Bioactive compounds and their interactions with EBCAs antagonistic to BW and PPN.
Regarding the use of bioactive compounds patners agreed to use chitosan as bioactive compound with potential as modulator of EBCAs against BW and PPN. In this task, UA implemented a system in which treatments with chitosan irrigation were included. Banana plants in pots with sterile peat were irrigated during 30 days with two concentrations of chitosan (0.1 [T1] and 1 mg ml [T2]) based on results obtained on tomato, barley and Arabidopsis. After 30 days plants were harvested and the growth parameters (root, shoot and corm weight and length) measured.
Results indicate that banana plants are tolerant to chitosan. Banana plants irrigated with a high concentration of chitosan (> 1mg ml) did not show significant differences respect to untreated controls. Chitosan irrigation at this concentration reduces ca. 50% growth of tomato plants.
UA team is currently designing RNAseq experiments to determine gene expression of Banana plants exposed treatments with combination of Pc and chitosan in Magenta box system, to characterise activation of banana defence genes during early events of Pc root colonisation, when the fungus is growing with and without chitosan. These experiments with the following treatments were designed:
- banana plants in contact with 21 day-old Pc colony on CMA in magenta box 5 days.
- banana plants in contact with 21 day-old Pc colony on CMA amended with chitosan (1mg ml) in magenta box 5 days.
- banana plants in an uninoculated magenta box with CMA for 5 days.
- banana plants in an uninoculated magenta box with CMA amended with chitosan for 5 days.
- magenta box with cellophane of Pc 21 day-old colonies on CMA.
- magenta box with cellophane of Pc 21 day-old colonies on CMA amended with chitosan.
Last two treatments allow extraction of good quality RNA of fungus only as RNAseq control.
UA team is also applying metabolomics to analyse banana plant rhizodeposition. In vitro plants (task 3.2) were placed in SDW overnight in the dark (24ºC, 65% humidity). Rhizodeposition was analysed using fluorescence, NMR and HPLC. A preliminary EEM fluorescence spectrum was produced.
Fluorecence analysis of banana root exudates generate after PARAFAC principal component analysis two components (C1 and C2). C1 (290-434 nm, 345-434 nm) corresponding to high molecular weight phenolics (fulvic and humic acids type), C2 (230-336nm, 280-336 nm) corresponding to low molecular weight phenolics (amino acids and peptides). NMR and HPLC analyses are in progress.
A number of experiments are under course at CENSA for the identification and evaluation of bioactive compounds with an impact on BW (subtask 3.3.1) or PPN (subtask 3.3.2.). For the latter, metabolites produced by fungi such asAcrostalagmusluteoalbusare under evaluation. An assay has been carried out on the effect of filtrates produced by a number of Trichodermaspp. proceeding from Algeria, in collaboration with ENSA (prof. S. Sellami). Filtrates of two isolates tested in vitroshowed a negative effect on juveniles of Meloidogyne incognita. A further assay has been carried out by in vitroby CNR using trimethylxhantine (caffeine) as bioactive compound. Exposure of M. incognitaeggs at a concentration of 4000 ppm completely inhibited eggs hatching after three weeks, although the eggs could complete their embryonic development. A bacteriostatic effect was also observed in vitroon egg-associated bacteria.
Task. 3.4 Risk analysis in the banana and ensete defence response.
To determine the effect of P.chlamydosporiaas an enhancer of plant defences UA studied how the endophytic fungus colonises banana plants in a time-course (1-7 days), to determine by qRT-PCR the transcriptomical response of the fungus during early plant colonisation. In addition, UA analysed how this fungus overexpresses genes related with activation of defence response, mainly genes involve in stress hormones homeostasis (like salicylic and jasmonic acid).
Distribution and quantification of Pochonia chlamydosporiain banana roots.Root culturing was used to evaluate the inoculation of P. chlamydosporia(e.g. detecting the fungus) in banana roots. P. chlamydosporiatotal and endophytic root colonization were analysed. The spatial pattern of root colonization by Pc123gfp was assessed using laser-scanning confocal microscopy. Ten fragments/root system (5 to 10 mm long) were examined in a Leica DM IRBE2 confocal microscope. GFP fluorescence and root cell autofluorescence were detected. Further detection of P. chlamydosporiain roots was performed by PCR. For this purpose, gDNA was extracted from roots of 10 plantlets/treatment (non-sterilized or surface-sterilized). Primers for PCR amplification of P. chlamydosporia were derived from Pc SCP1 and Permease1 genes.
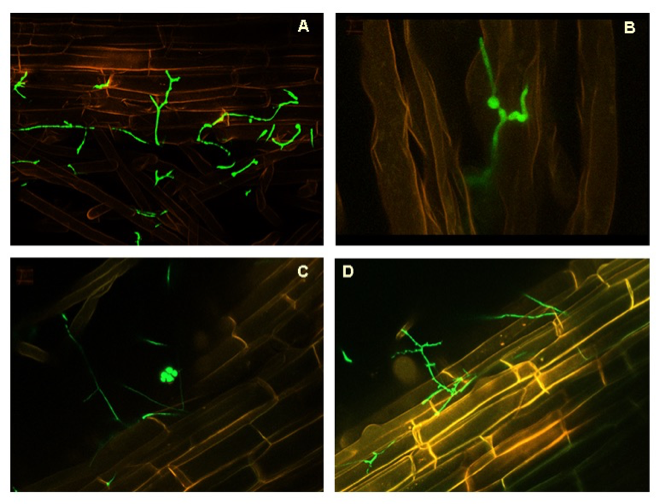
Magenta-box experiments (System 1). UA evaluated endophytic colonisation of banana plants by P. chlamydosporiausing molecular and physiological analyses to enhance banana plant defense response. Banana roots were inoculated with Pc123 isolate transformed with the green fluorescent protein (GFP). UA showed the Pc colonisation of the rhizoplane, root hairs (Fig 6 A,B). However, the fungus also colonised endophytically root cells (Fig. 6 C, D).
Quantification of Pc colonization in banana plants by culturing methos.The UA axenic systems allowed external colonisation of banana roots. The fungus increased endophytic (internal) colonisation of roots 1.6 fold in 7 days.
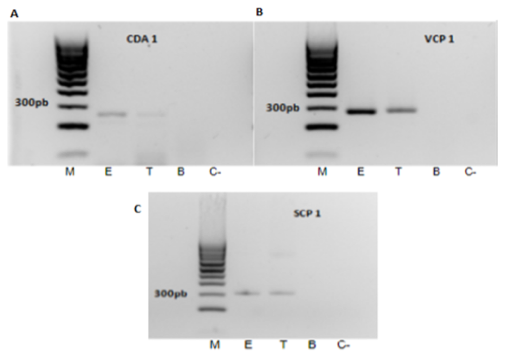
Molecular identification of Pc colonising banana roots endophytically.Pc SCP1 clearly reported both endophytic and total root colonisation by Pc (Fig. 7 A, ca. 400 pb band), confirming the amplicon identity by sequencing. Pc permease 1 gene displayed no PCR aspecificities but was less sensitive to detect endophytic root colonisation by the fungus (Fig. 7 B, ca. 250 bp).
Colonization of banana roots by P. chlamydosporiaafter 5d in contact with the fungus was also measured using quantitative PCR, measuring the amout of fungal DNA by evaluating VCP1 gene with specific primers. Banana DNA quntification was performed by amplification of PR1 gene using specific primers. Serial dilutions of isolate Pc123 genomic DNA defined a calibration curve, using three independent calibrations for each DNA sample. After each run, a dissociation curve was acquired to check for amplification specificity. PCR data confirmed that endophytic colonisation of banana roots by P. chlamydosporiais lower (ca. 5 fold less) than total root colonisation.
To characterize transcriptomic response of P. chlamydosporiaduring banana root colonisation, UA evaluated expression by qRT-PCR of Pc genes after 5 days in contact with banana root in a Magenta, testing genes related to PPN egg infection (SCP1, VCP1 proteases) chitin/chitosan metabolism (chitin deacetylases and chitosanases) and also genes encoding proteins putatively involved in modulation of plant immunity (LysM effectors). The fungus expressed those genes when colonizing banana plants endophyticaly. Also the plants showed expression of PR1 and PR3 when fungus is colonizing the rhizosphere (Fig. 62).
UA team is currently designing RNAseq experiments to determine the whole gene expression of banana plants exposed to P. chlamydosporiain a time-course (1 -7 days) in the magenta box system, to characterise activation of defense genes during early events of root colonisation. This experiment was designed with three treatments:
- banana plants in contact with 21 day-old P. chlamydosporiacolony in magenta box for 2, 5 and 7 days;
- banana plants in a uninoculated magenta box for 2, 5 and 7 days;
- cellophanes of P. chlamydosporia21 day-old colonies (see RNAseq in task 3.3).


